
NMsim - Seamless NONMEM Simulation Platform in R
Philip Delff
February 20, 2026 using NMsim 0.2.6.902
Source:vignettes/NMsim-intro.Rmd
NMsim-intro.RmdObjectives
This introduction to NMsim aims at enabling NONMEM users to
- Configure NMsim to find your NONMEM installation
- Set up a simulation data set and simulate a NONMEM model using that data set
- Simulate a typical subject
- Simulate multiple models and compare them
- Simulate observed or previously simulated subjects based on emperical Bayes estimates (ETA’s)
- Simulate multiple subjects with covariate sampling and generation of prediction intervals
Configuration
NMsim must be configured with the path to the NONMEM executable. This
can be done for each NMsim() call using the
path.nonmem argument, but more easily it can be configured
globally the following way. Also including where NMsim will run NONMEM
and store intermediate files (dir.sims) and where to store
final results (dir.res).
library(NMdata)
## Point NMsim to your NONMEM exectuable - looks like this on linux/osx
NMdataConf(path.nonmem = "/opt/NONMEM/nm75/run/nmfe75")
## or on Windows, it could be
NMdataConf(path.nonmem = "c:/nm75g64/run/nmfe75.bat")
NMdataConf(dir.sims="simtmp-intro", ## location of sim tmp files
dir.res="simres-intro") ## location of sim resultsFor more information on this and how to test the configuration, see
NMsim-config.html.
A First Simulation with NMsim()
When providing a simulation data set, the default
NMsim() behavior is to sample a new subject (ETA’s).
## Point to the model to estimate
file.mod <- system.file("examples/nonmem/xgxr021.mod",
package="NMsim")
## Easily create a muliple-dose simulation data set with a loading dose
data.sim <- NMcreateDoses(TIME=c(0,24),AMT=c(300,150),ADDL=5,II=24,CMT=1)|>
NMaddSamples(TIME=0:(24*7),CMT=2)
## Simulate
simres <- NMsim(file.mod=file.mod,data=data.sim,table.vars=c("PRED","IPRED","Y"))The simulation input data set is a data.frame, and
NMsim() returns a data.frame. Plot of simulation results
(click to button show code):
datl <- as.data.table(simres) |>
melt(measure.vars=c("PRED","IPRED","Y"))
plot1 <- ggplot(datl,aes(TIME,value,colour=variable))+
geom_line(data=function(x)x[variable!="Y"])+
geom_point(data=function(x)x[variable=="Y"])+
labs(x="Hours since first dose",y="Concentration (ng/mL)",
subtitle="Simulation of one new subject.",
colour="")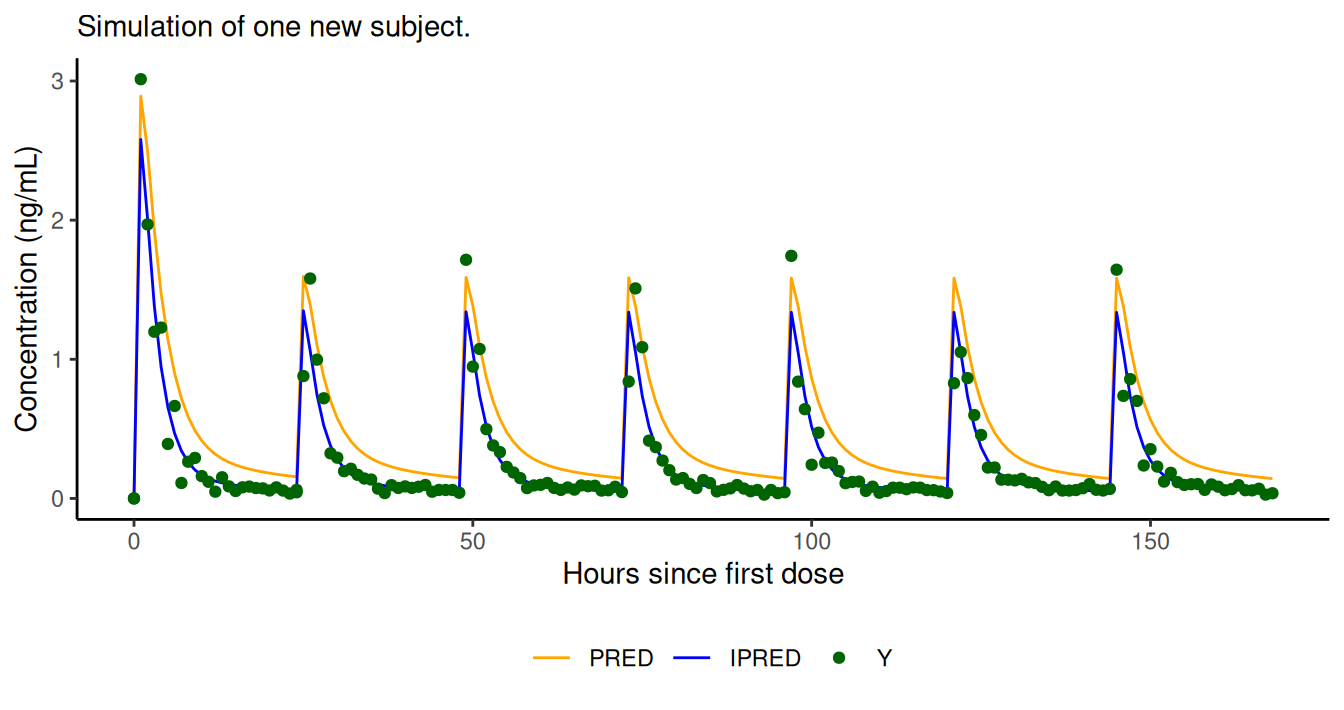
PRED, IPRED, and Y (if defined in
control stream) are easily obtained with NMsim.
Notice that for running a model simulation, no information about the
model is needed except for the control stream file path. The simulation
is based on evaluation of PRED, IPRED, and
optionally Y. Options exist for building more advanced
simulation models. The models shown here are based on data available in
the xgxr
package.
Here is a conceptual diagram of the NMsim() workflow.
file.mod: Existing file.mod (unmodified NONMEM control stream)
data: A data.frame with simulation data
│
▼
NMsim()
│
├─► Creates simulation control stream in dir.sims/
│ (Typical automatic modifications:
│ adds $SIMULATION, comments out $ESTIMATION,
│ modifies data sections)
│
├─► Runs NONMEM in dir.sims/
│ (raw NONMEM outputs: lst, tab, phi, etc.)
│
├─► Copies/organizes results into dir.res/
│ ..._MetaData.rds: Main results file for re-rereading results
│ ..._ResultsData.fst: Compressed archive
│
└─► Reads results → returns to R
Diagram of the NMsim() process. The user calls NMsim() on an existing control stream, typically developed for estimation and a simulation data set as a data.frame (NMsim will store as file for NONMEM). Simulatio results are returned to R.Before getting into details on how to create datasets and other features, let’s nail down some properties of NMsim to avoid common misconceptions. As you start to explore NMsim, remember:
- NMsim automates simulation of Nonmem models. You never have to edit
the Nonmem control stream manually. The path to a control stream
(typically of an estimated model) is passed to NMsim, and NMsim will
then internally create a simulation control stream, execute it, and
return the results.
- You should never save the simulation data set in a text file (like
csv or similar). Just pass the data.frame in the
dataargument inNMsim(), and NMsim will handle and save the data internally, including needed modifications to the simulation control stream to use the provided data.
- Re-read 1. This also means you never have to modify the control
stream to match your simulation data set. NMsim fully automates
this.
- You do not have to manually read simulation output tables. NMsim()
reads those and returns them in a data.frame by default. When you do
simres <- NMsim(file.mod=my_file_path,data=my_df),simrescontains (the combination of) the output table(s).
- Re-read 1. You do not have to update the control stream with final
parameter estimates. Those are by default inserted for simulation by
NMsim().NMsim()obtains them from the.extfile associated with the provided control stream.
List of NMsim Modifications to Control Stream
To illustrate what NMsim did to the provided control stream, the
following uses waldo::compare() to compare the provided
file.mod and the generated simulation control stream.
Notice that $DATA and $INPUT have been updated
to match the provided simulation input data, parameter sections (in this
case THETA and OMEGA) updated with final
parameter estimates. Estimation and covariance steps have been replaced
by a simulation step, and $TABLE modified to generate a new
output table.
## file.mod$INPUT vs simulation$INPUT
## - "$INPUT ROW ID NOMTIME TIME EVID CMT AMT DV FLAG STUDY"
## - "BLQ CYCLE DOSE PART PROFDAY PROFTIME WEIGHTB eff0"
## + "$INPUT NMROW ID TIME EVID CMT AMT II ADDL DV MDV"
##
## file.mod$DATA vs simulation$DATA
## - "$DATA ../data/xgxr2.csv IGNORE=@ IGNORE=(FLAG.NE.0) IGNORE(DOSE.LT.30)"
## + "$DATA NMsimData_xgxr021_noname.csv"
## + "IGN=@"
##
## file.mod$THETA | simulation$THETA
## [1] "$THETA (0,1) ; POPKA" - "$THETA (0,2.16656) ; POPKA" [1]
## [2] "$THETA (0,100) ; POPV2" - "$THETA (0,75.7289) ; POPV2" [2]
## [3] "$THETA (0,3) ; POPCL" - "$THETA (0,13.9777) ; POPCL" [3]
## [4] "$THETA (0,50) ; POPV3" - "$THETA (0,150.06) ; POPV3" [4]
## [5] "$THETA (0,.5) ; TVQ" - "$THETA (0,8.48651) ; TVQ" [5]
##
## file.mod$OMEGA | simulation$OMEGA
## [1] "$OMEGA 0 FIX ; BSV KA" - "$OMEGA 0 FIX ; BSV KA" [1]
## [2] "$OMEGA 0.1 ; BSV V2" - "$OMEGA 0.178666 ; BSV V2" [2]
## [3] "$OMEGA 0.1 ; BSV CL" - "$OMEGA 0.249779 ; BSV CL" [3]
## [4] "$OMEGA 0 FIX ; BSV V3" - "$OMEGA 0 FIX ; BSV V3" [4]
## [5] "$OMEGA 0 FIX ; BSV Q" - "$OMEGA 0 FIX ; BSV Q" [5]
##
## file.mod$TABLE vs simulation$TABLE
## - "$TABLE TVKA TVV2 TVV3 TVCL KA V2 V3 CL Q PRED IPRED Y NOPRINT FILE=xgxr021_res.txt"
## + "$TABLE NMROW PRED IPRED Y NOPRINT NOAPPEND ONEHEADERALL NOTITLE FORMAT=s1PE16.9 FILE=xgxr021_noname.tab"
##
## `file.mod$ESTIMATION` is a character vector ('$ESTIMATION METHOD=1 POSTHOC INTER MAXEVAL=9999 NSIG=2 SIGL=9', ' PRINT=10 NOABORT MSFO=xgxr021.msf')
## `simulation$ESTIMATION` is NULL
##
## `file.mod$SIMULATION` is NULL
## `simulation$SIMULATION` is a character vector ('$SIMULATION ONLYSIM (2134995937) ')Simulation Data Sets
The input data is just a data.frame. NMsim will save the
data set and create
- It must contain at least the variables NONMEM will need to run the
model (typically
ID,CMT,AMT, etc. plus covariates) - It can contain character variables (automatically carried to results)
- Column order does not matter
As long as those requirements are met, there are no requirements to
how the data sets are created. So if you already have a prefered way to
do this, that’s fine. NMsim provides convenient helper functions that
can optionally be used. NMcreateDoses() creates just the
dosing events, and NMaddSamples() adds the sampling events.
By default, doses are indicated using EVID=1 and samples by
EVID=2.
doses <- NMcreateDoses(TIME=seq(0,by=24,length.out=7),
AMT=c(300,150),CMT=1)NMcreateDoses() offers arguments matching the
NONMEM-specific variables such as CMT, EVID,
ADDL, II, SS and more. This
example uses ADDL and II to repeat the
maintenance dose. It is designed to offer convenient syntax for most
common dosing regimens.
Adding Sampling Times
For adding sampling times to a set of doses and/or samples,
NMaddSamples() provides a similarly flexible interface. It
accepts data.frames with covariates allowing for different sampling
schemes for different subject groups (say different dosing regimens),
and dosing times can be supplied relative to previous dosing times.
dat.sim <- NMaddSamples(doses,TIME=0:(24*7),CMT=2)NMaddSamples() has many useful features, including a
TAPD argument for adding samples relative to previous
dosing time, adding sampling for multiple endpoints, and different
sampling times for different subgroups of subjects. See Data Set Creation with
NMsim for more details on creation of data sets, including
use of NMcreateDoses() and NMaddSamples() and
how to also add time after previous dose and more.
Check The Simulation Dataset
A quick way to check for a lot of common issues in a NONMEM data set
is running NMcheckData():
NMcheckData(dat.sim,type.data="sim")It is also advised to plot the simulation data set. See Creation of Simulation Data Sets for more details.
Typical Subject Simulation
- A typical subject is a subject with all ETAs = 0
- Covariates values are supplied using the simulation input data set
-
typical=TRUE: replace all$OMEGAvalues with zeros
simres.typ <- NMsim(file.mod=file.mod,data=data.sim,
typical=TRUE, ## FIX all OMEGA's to zero
name.sim="typical", ## simulation name - included in output
)Simulate Multiple Models
Multiple models can be simulated using the same data set in one
function call by supplying more than one model in the
file.mod argument. The models can be simulated on multiple
data sets by submitting a list of data.frames in the data
argument. NMsim will return one data.frame with all the results for easy
post-processing.
file2.mod <- "models/xgxr114.mod"
simres.typ2 <- NMsim(file.mod=c("2 compartments"=file.mod,
"1 compartment"=file2.mod),
data=data.sim,
typical=TRUE, ## FIX all OMEGA's to zero
table.vars=c("PRED","IPRED","Y")
)
## The "model" column is used to distinguish the two models
subset(simres.typ2,EVID==2) |>
ggplot(aes(TIME,PRED,colour=model))+
geom_line()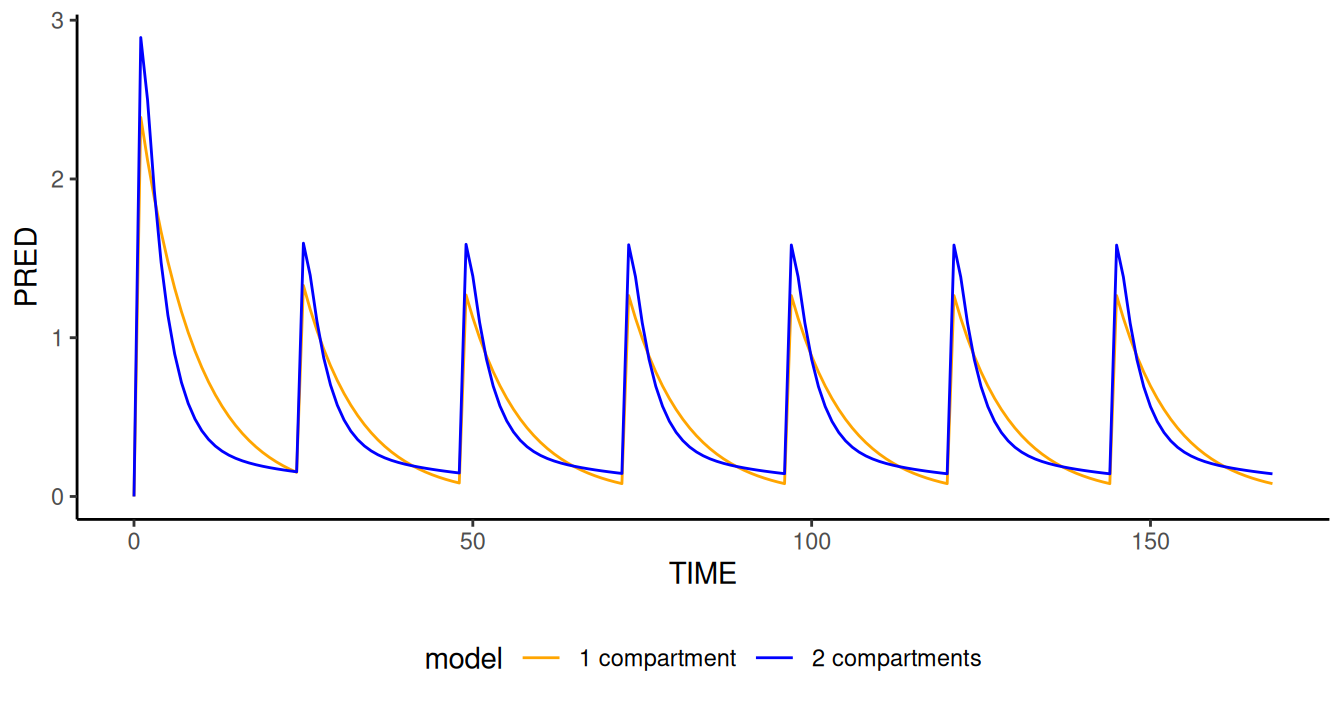
Simulation of multiple models and even multiple data sets is handled
within one NMsim() call.
Emperical Bayes’ Estimates (known ETAs)
Reusing ETA’s is enabled using the NMsim_EBE method.
- By default, automatically re-uses estimated individual ETAs
- ID values in simulation data must match the ID values in the estimation that you want to simulate
- Other ETA sources (
.phifiles) can be specified - Does not simulate residual variability - see
addResVar()if needed - Remember: Covariates may be needed in data set to fully reproduce the subjects’ parameters
In the following, we use table.vars to specify variables
to output in NONMEM’s $TABLE section. In this case, we do
that to make sure we get CL and V2. But
generally, table.vars is very important to know as the very
first thing to do to speed up NMsim(). This is because
NONMEM often takes much longer writing the output table than it does
doing the actual simulation. So it is recommended to specify a slim
output table using something like
table.vars=c("PRED","IPRED","Y") and other variables you
may need from NONMEM. Notice NMsim knows how to combine
output table data with the simulation input data, so you do not need
variables like ID or TIME in
table.vars.
## this example uses the same sim data for all subjects
res <- NMscanData(file.mod,quiet=T)
ids <- unique(res$ID)[1:5]
data.sim.ind <- merge(subset(data.sim,select=-ID),
data.frame(ID=ids))
setorder(data.sim.ind,ID,TIME,EVID)
simres.ebe <- NMsim(file.mod,
data=data.sim.ind,
table.vars=c("CL","V2","IPRED","PRED")
)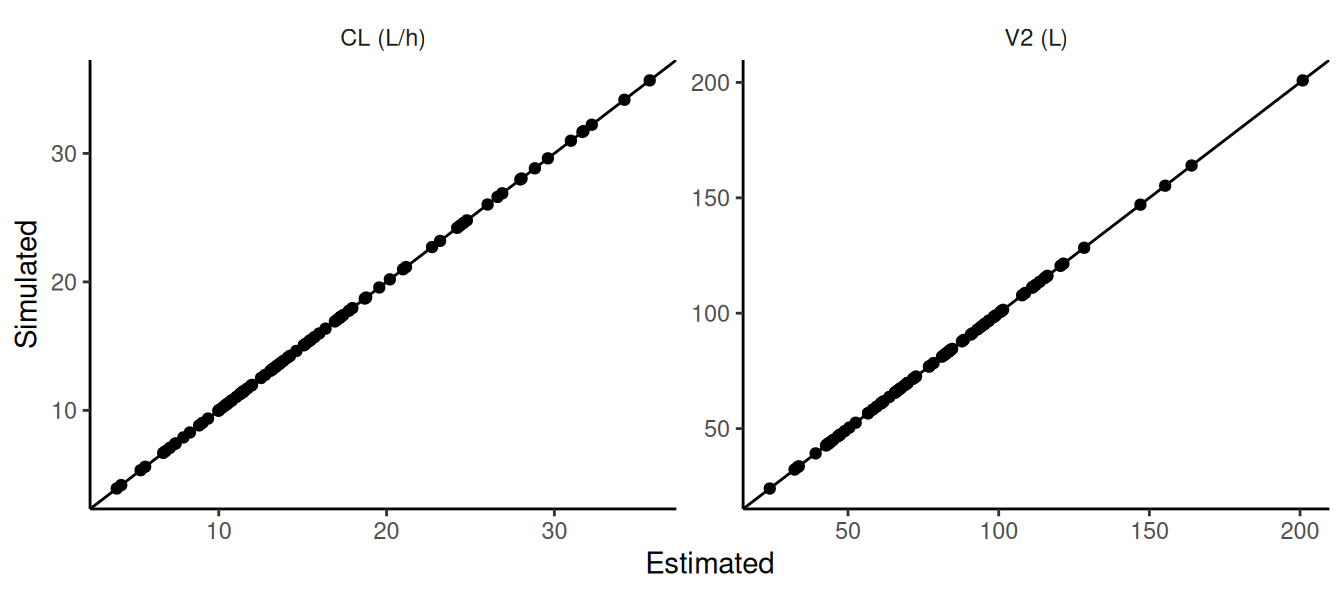
Individual parameters are confirmed to be identical in estimation results and simulation results
Prediction Intervals
New subjects can be simulated in multiple ways with NMsim.
- If the input data set contains multiple subjects, these subjects
will get separate random effects due to NONMEM
$SIMULATION - The
subproblemsargument translates to theSUBPROBLEMSNONMEM subroutine, replicating the simulation the specified number of times with new seeds - The
simPopEtas()function can generate a synthetic .phi file with a simulated population that can be reused in futureNMsim()calls. This can be combined with simulation of covariates in R, allowing reuse of the same subjects across multiple simulations.
In the following we use both of these approaches to simulate 1000 new
subjects. We use NMsim()’s name.sim argument
to distinguish the simulation in output data and simulation output
files.
- Simulate 1000 new subjects using
$SUBPROBLEMS
Notice the following reuses the input data set 1000 times, and a
column called NMREP will count the subproblem number in the
output.
tablevars=cc(PRED,IPRED,Y)
simres.subprob <- NMsim(file.mod=file.mod,
data=data.sim,
name.sim="Subproblems", ## naming the simulation
subproblems=1000, ## Will become SUPROBLEMS=1000 in NONMEM
table.vars=tablevars,
seed.R=764, ## NMsim() will set the R seed for reproducibility
reuse.results=reuse.results
)To generate a prediction interval, this format is sufficient. If you
want to distinguish the subjects, you can update the ID
column to reflect unique combinations of ID and
NMREP. With data.table, this can be done this
way:
## data.table:
as.data.table(simres.subprob)[,ID:=.GRP,by=.(NMREP,ID)]
## dplyr:
simres.subprob <- simres.subprob |>
group_by(NMREP,ID) |>
mutate(ID = cur_group_id()) |>
ungroup()By the way, if you would like NMsim() to return
data.table objects, just run
NMdataConf(as.fun="data.table"). If you want tibbles, run
NMdataConf(as.fun=tibble::as_tibble).
- Simulate 1000 new subjects with covariate sampling
The NMsim package provides a convenient funtion to sample covariates
from an existing data set. We have a simulation data set with only one
subject. We want to replicate that data set to create 1000 subjects with
covariates sampled with replacement from the analysis data set.
sampleCovs() does exactly that. Notice two important
features of sampleCovs():
It automatically makes sure to sample with equal probability from all subjects, independently of how many data points they have in the analysis set.
For a simulated subject, all covariates are sampled from the same subject in the analysis set. This ensures preservaion of the correlation of the covariates from the analysis set to the simulation.
## read the analysis data set from the model we are simulating from
dt.covs <- NMscanData(file.mod,quiet=T)
## replicate the simulation data set (one subject) to create 1000
## subjects with covariates "WEIGHTB" and "trtact" sampled from the
## analysis set.
data.sim.nsubjs <- sampleCovs(data=data.sim,data.covs=dt.covs,
covs=c("WEIGHTB","trtact"),
Nsubjs=1000,seed.R=2372)
## renaming trtact to shorter trt in this example
setnames(data.sim.nsubjs,"trtact","trt")
## generate the population first, by simulating etas to use in the sim
simPopEtas(file.mod=file.mod,N=1000,seed=1231,
file.phi="simres-intro/xgxr021_1000subjs.phi")
simres.datarep <- NMsim(file.mod=file.mod,
data=data.sim.nsubjs,
name.sim="Individual simulation data",
table.vars=tablevars,
seed.nm=103,
method.sim=NMsim_EBE,
file.phi="simres-intro/xgxr021_1000subjs.phi",
reuse.results=reuse.results
)- Derive and plot 90% prediction intervals
## Collect and stack simulation results
simres.newpops <- rbind(as.data.table(simres.subprob),
simres.datarep,fill=T)[EVID==2]
## Derive prediction intervals - notice name.sim distincts results from the two methods
simres.pi <- simres.newpops[
,setNames(as.list(quantile(IPRED,probs=c(.05,.5,.95))),cc(ll,median,ul)),
by=.(name.sim,trt,TIME)]
label.pi <- "90% Prediction interval"
simres.pi$type <- label.pi
p.pi <- ggplot(simres.pi,aes(TIME,fill=type))+
geom_ribbon(aes(ymin=ll,ymax=ul),alpha=.4)+
geom_line(aes(y=median,linetype="Median"))+
scale_alpha_manual(values=setNames(c(.5),label.pi))+
scale_linetype_manual(values=setNames(c(1),"Median"))+
facet_wrap(~name.sim)+
labs(x="Hours since first dose",y="Concentration (ng/mL)",colour="",linetype="")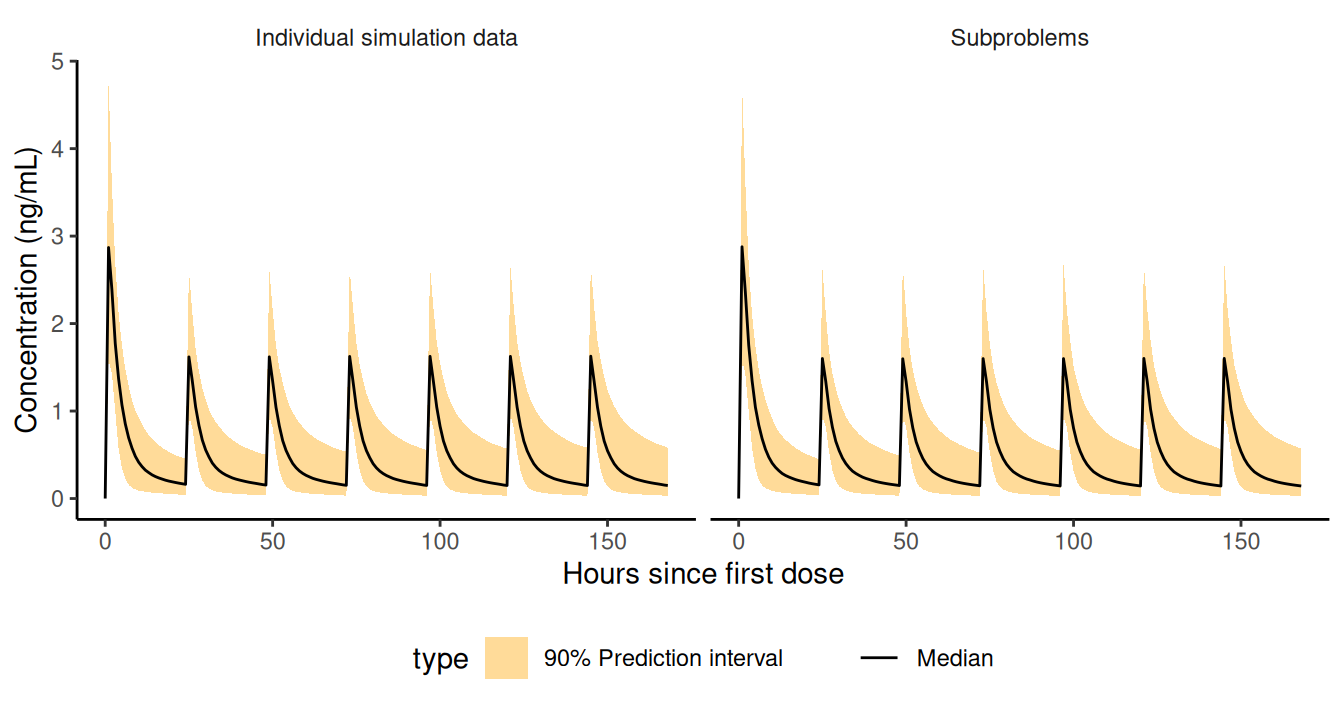
Prediction intervals. New subjects can be simulated in multiple ways with NMsim. A simulated population can be reused across simulations.
Read Previously Generated Simulations
There is no need to save simulation results because they are already
saved by NMsim. Instead, use arguments
dir.sims, dir.res and name.sim to
make sure to get a meaningful structure for the generated files. Then
read the results with NMreadSim(). To re-read the first
simulation we did in this article, we can do this:
simres <- NMreadSim("simres-intro/xgxr021_noname_MetaData.rds")The folder and file names were constructed based on
dir.res="simres-intro" and because name.sim
was not provided for that first simulation, in which case “noname” is
used as a placeholder. In fact, if we look at the console output from
NMsim, it is telling us exactly that (look at the last line).
Click to show R console output from NMsim
> simres <- NMsim(file.mod=file.mod,data=data.sim)
Location(s) of intermediate files and Nonmem execution:
simtmp-intro/xgxr021_noname
Location of final result files:
simres-intro
* Writing simulation control stream(s) and simulation data set(s)
* Executing Nonmem job(s)
Starting NMTRAN
(...)
Done with nonmem execution
* Collecting Nonmem results
Simulation results returned. Re-read them without re-simulating using:
simres <- NMreadSim("simres-intro/xgxr021_noname_MetaData.rds")